Summary

During my research career, I have worked and am working on several data science-related projects. Applications vary from separation science, through pharmaceutical chemistry to proteomics and chemical engineering. The works feature exploratory data analysis (principal component analysis, partial least squares), feature selection (genetic algorithms, particle swarm optimization, firefly / flower pollination algorithms, LASSO, LARS), mathematical programming/optimization (single- and multi-objective optimization, non-linear programming), (high-dimensional) modelling (multiple linear regression, partial least squares, discriminant analysis – linear/quadratic, hierarchic cluster analysis), machine learning techniques (artificial neural networks, convolutional neural networks, random forests, gradient / adaptive boosting, CART, support vector machines, genetic programming) among others. The following section represents a summary of my on-going and completed research with main deliverables. For a list of publications please refer to my Google Scholar profile or the Publications page on this website.
Machine learning for solubility prediction
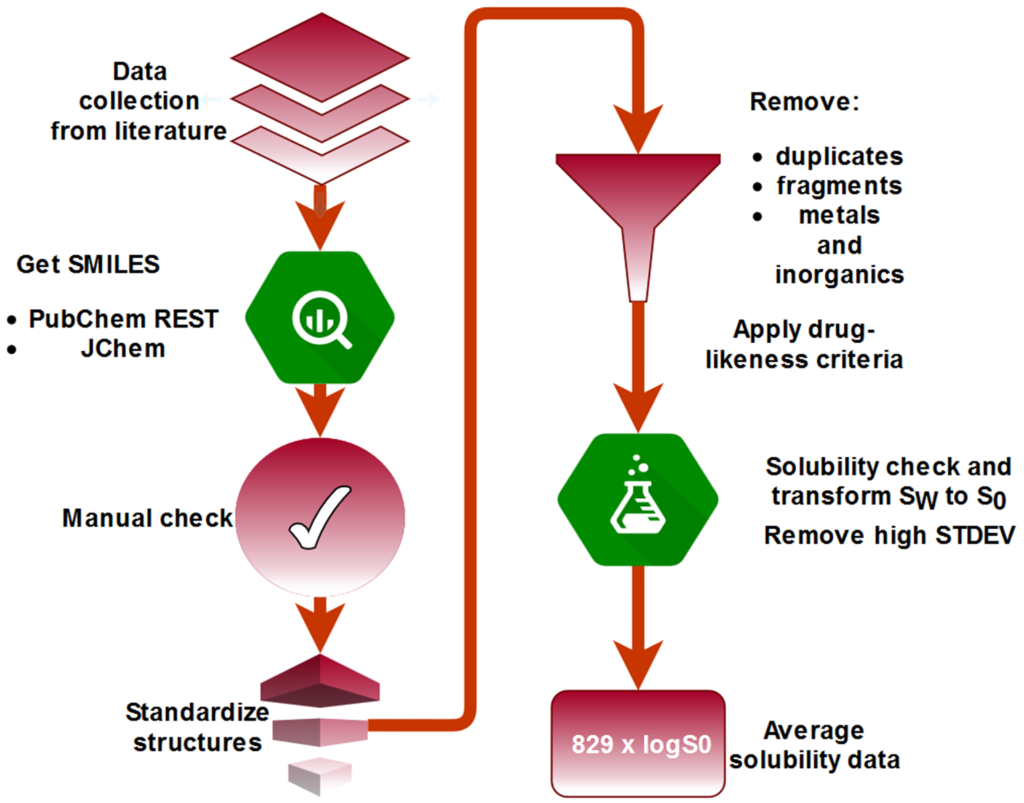
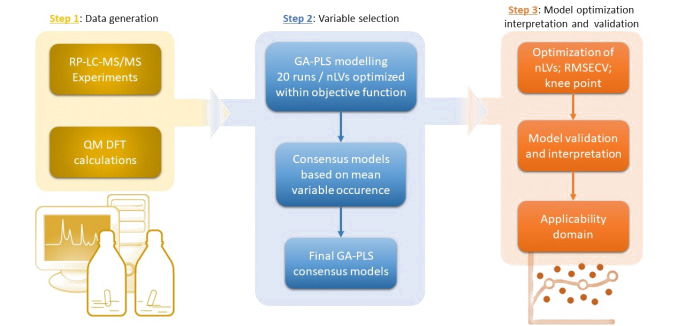
QSPR models were developed for a set of publicly available intrinsic solubility data of 829 drug-like compounds. Four different machine learning algorithms (Random forest, LightGBM, Partial least squares and LASSO) coupled with multi-stage permutation importance for feature selection and Bayesian hyper-parameter optimization were employed for prediction of solubility based on chemical structural information. Our results have shown that LASSO yielded the best predictive ability on an external test set. Taking into account a the number of descriptors besides RMSE(test), an RF model achieved the best balance between complexity and predictive ability (in progress).
QSRR of flavonoids
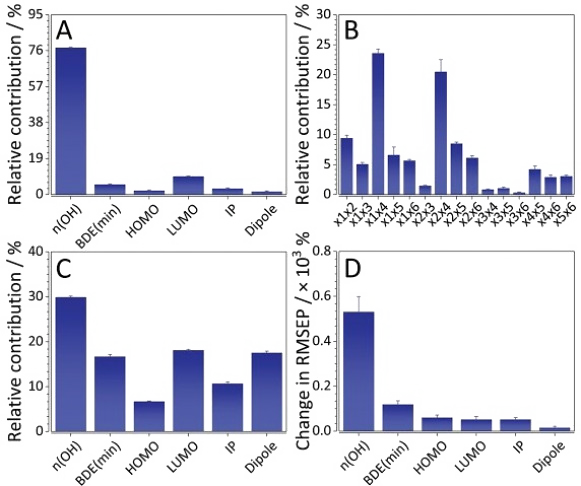
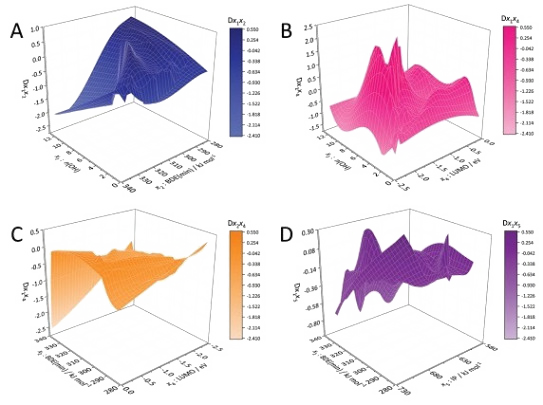
Development of quantitative structure-retention relationships (QSRRs) models for a series of flavonoids on three chromatographic column (C18, F5 and IAM). Quantum mechanical parameters are calculated and correlated with retention time, mechanistic interpretations are given for all the columns. Parameters of the first step of the HAT and SPLET mechanisms (of antioxidant activity) considered as predictors. Solvation energy, number of OH groups, most negative charge, dipole moment, HOMO, LUMO, HOMO-LUMO gap, softness, hardness, chemical potential, electronegativity, and electrophilicity index also considered. (complete).
Elution order prediction
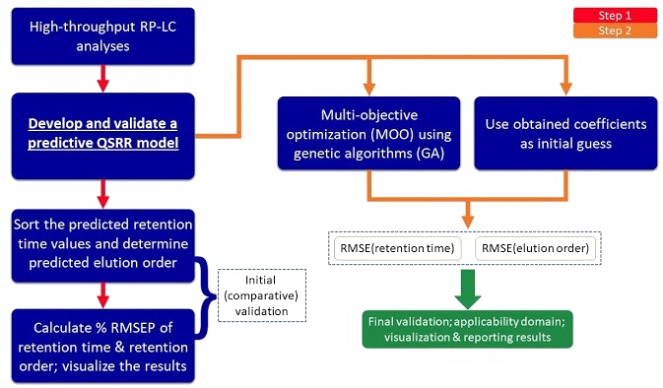
Prediction of chromatographic elution order from quantitative structure-retention relationships through mathematical programming (non-linear programming – NLP, multi-objective optimization – MOO) with relaxed constraints. Abbreviations (in order of appearance): RP-LC—reversed-phase liquid chromatography, RMSE—root mean square error, QSRR—quantitative structure-retention relationships. (complete, one paper published & one under review for publication, KR and US patents pending).
Biocrude reduction and optimization software
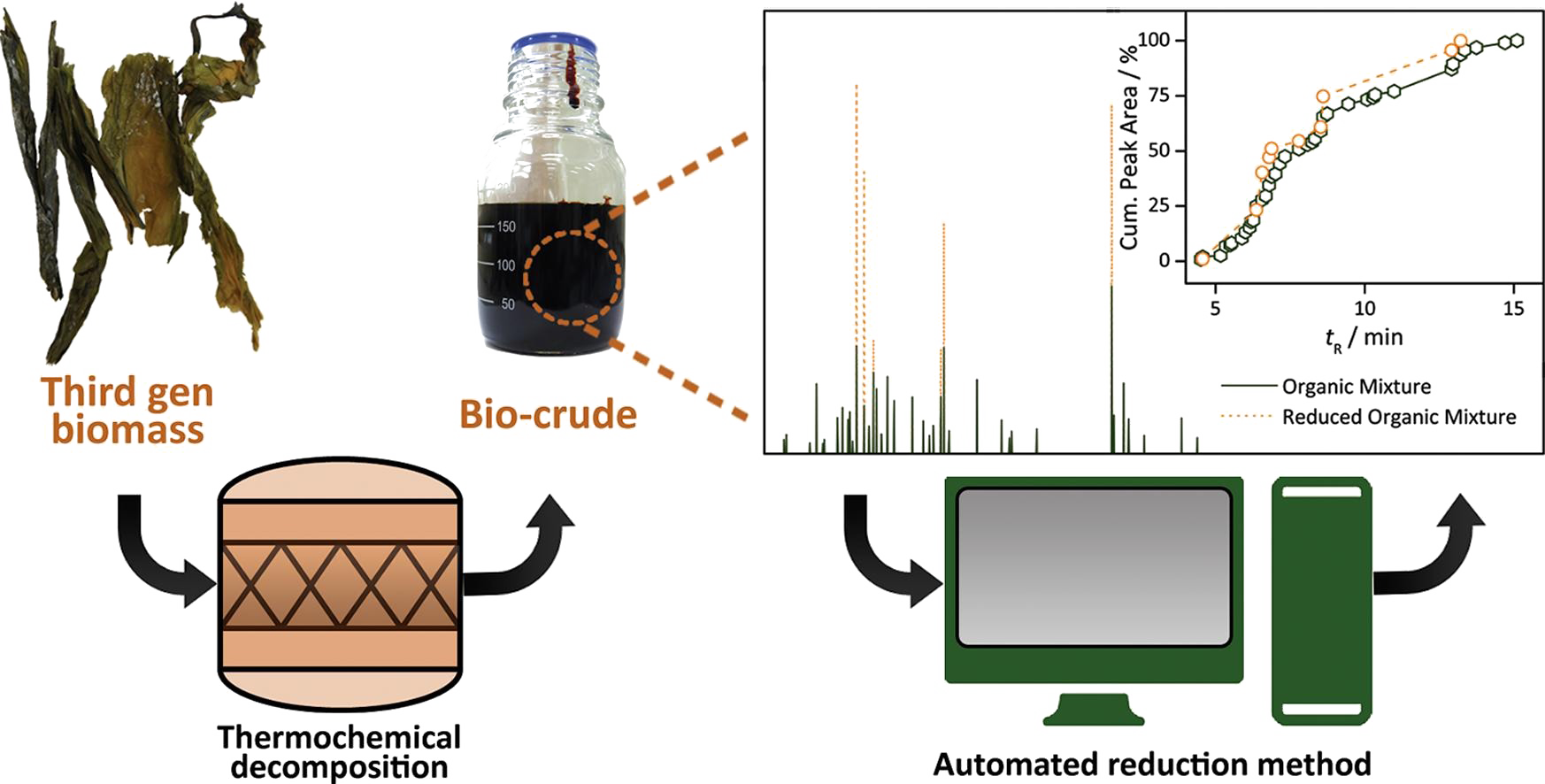
In this work, a software employing automated modelling of bio-crudes based on raw experimental data, has been developed. The output of this software is a ready-to-use reduced mixture, including all product phases and in mass balance with the Proximate and Ultimate analyses of the feedstock biomass material. As there are many approaches to bio-crude modelling, the novelty of this method lies in the combination of the minimization of the number of components needed and the minimization of the level of artificiality introduced in the system. The automation of the method allows for fast reduction and optimization. The biocrude reduction and optimization software (BROS) written entirely in Python and deployed on the web is a software for biocrude mixture modelling / reduction and optimization of mass and atomic balances. (Brigljevic, et al. Appl. Energy 2018, 215, 670-678) (project completed, software still in development, US and KR patents pending).
ANN-based QSAR models for prediction of antioxidant activity
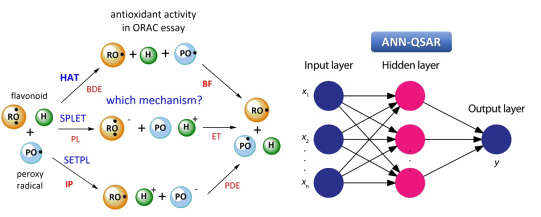
Development and optimization of an ANN-based QSAR model for prediction of anti-oxidant activity and elucidation of the mechanism of the ORAC assay (completed, publication in IJMS). Algorithms for analysis of contributions of input variables (e.g., molecular descriptors) towards the target variable(s) (e.g., anti-oxidant activity) in ANN models (completed, publication in JCC).